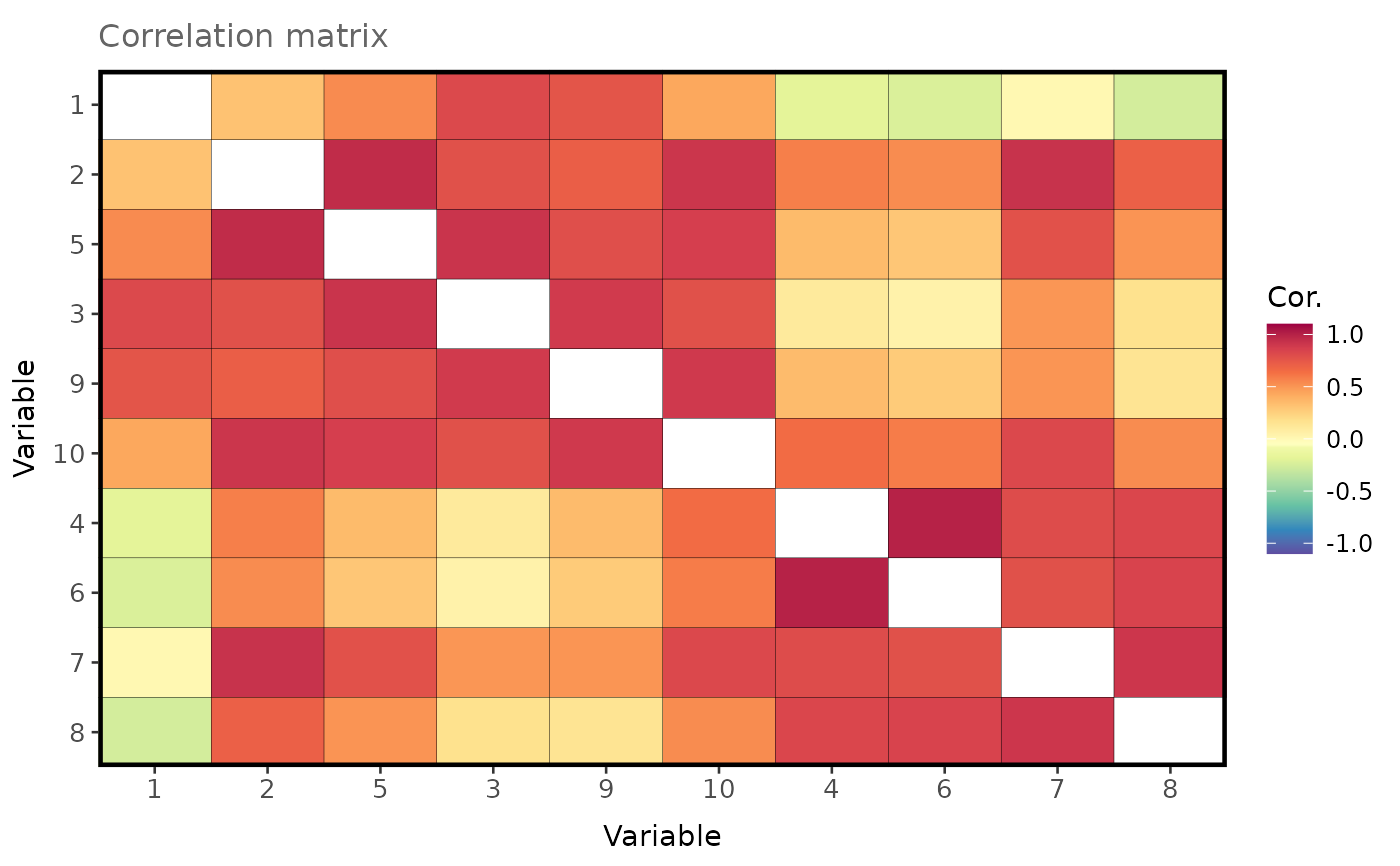
Simulate a structured correlation matrix with reduced rank
Source:R/random_correlation_matrix.R
struc_cor_mat.RdCreates a symmetric n x n correlation matrix with user-defined structure and rank.
Usage
struc_cor_mat(
n = 5,
base.cor = 0.5,
range = NULL,
rank = 3,
skew = 0,
base.mat = NULL,
pos.def = FALSE,
small.positive = NULL
)Arguments
- n
A scalar defining the dimensions of the correlation matrix.
- base.cor
A scalar defining the baseline correlation.
Note:-1 < base.cor < 1.- range
A scalar defining the range of correlations around the baseline. By default,
range = 1 - base.corwhich ensures the matrix is positive semi-definite with defined rank. Note:base.cor + range <= 1.- rank
A scalar defining the rank of the correlation matrix.
- skew
A scalar defining the skewness imposed on the correlations. Note:
-1 < skew < 1.- base.mat
An optional
n x nbase correlation matrix. When supplied,base.corandskeware ignored and noise is simulated based onrank.- pos.def
When
TRUE(default isFALSE), the functionbendof the R package `mbend` is used to bend a non-positive (semi)-definite matrix to be positive (semi)-definite.- small.positive
Argument passed to
bendwhenpos.def = TRUE(default is 1e-8). Eigenvalues smaller thansmall.positiveare replaced by this.
Note:0 < small.positive < 0.1.
Value
A symmetric n x n correlation matrix with defined rank.
When pos.def = TRUE, the correlation matrix is guaranteed to be positive (semi)-definite.
Examples
# Simulate and visualise a correlation matrix with 10 columns and rows, rank equal to 4 and
# negatively skewed correlations.
cor_mat <- struc_cor_mat(
n = 10,
base.cor = 0.3,
range = 0.7,
rank = 4,
skew = -0.5
)
plot_matrix(
mat = cor_mat,
order = TRUE
)